Part:BBa_K1998008
psbELJ-TB
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1286
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 67
Illegal AgeI site found at 173
Illegal AgeI site found at 457
Illegal AgeI site found at 466 - 1000COMPATIBLE WITH RFC[1000]
Overview
This composite part is composed of operons psbELJ and psbTB found in the photosystem II pathway of Chlamydomonas reinhardtii. Three subunits make up the psbELJ cluster and are involved in the structure of the thylkaoid membrane complexes. The psbT and psbB protein function is not well known however it appears to be involved in the dimerization of Photosystem II and is essential for photosynthetic activity.
The first operon encoded in this part contains psbE, psbL, and psbJ, driven by Plac promoter, with RBS preceeding each gene, and terminator at the 3' end. The second operon is composed of the psbT and psbB genes which both have a RBS preceding them.
This part contains two operons that make up our Photosystem II Pathway.
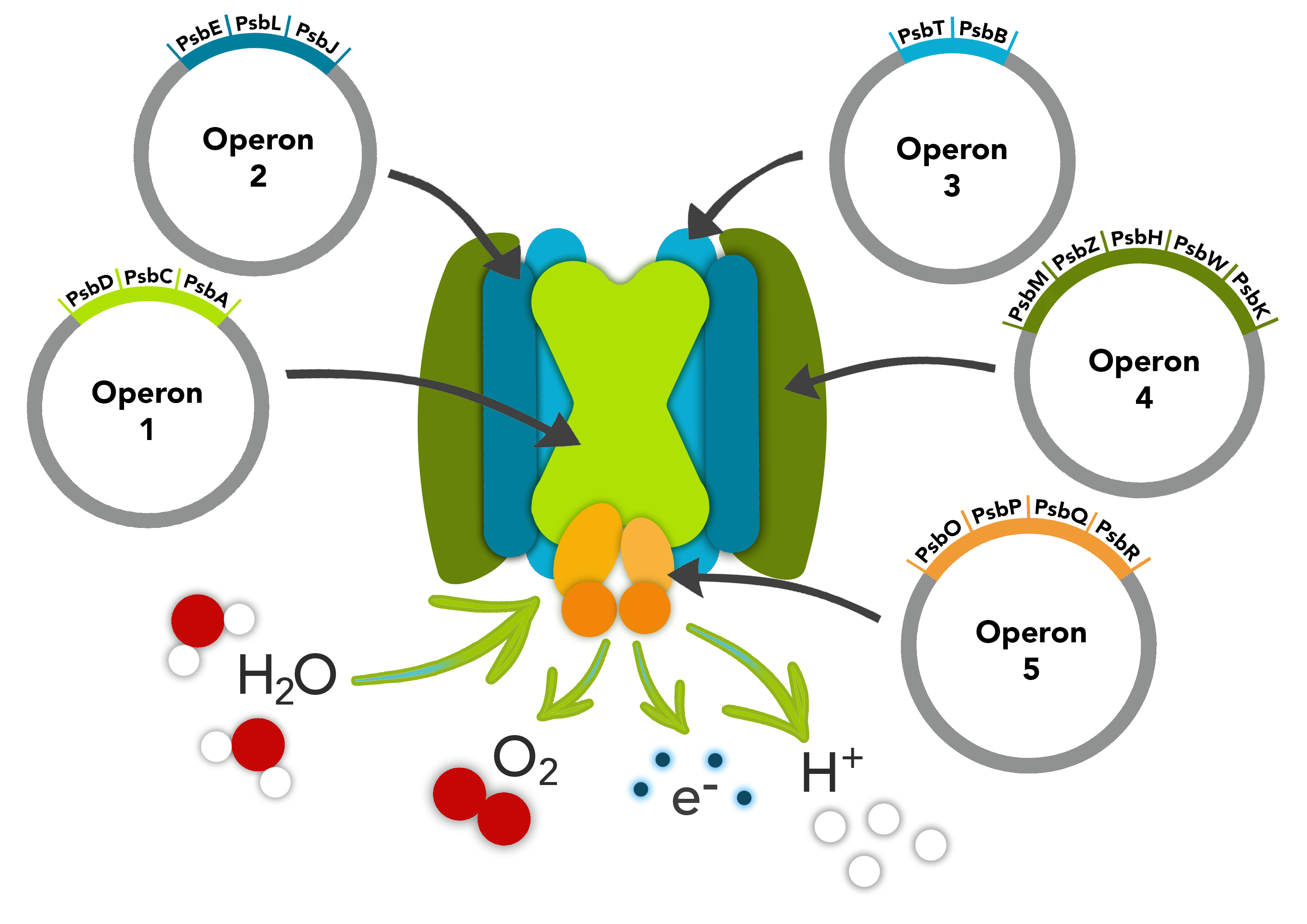
Biology & Literature
The first operon in this composite part is composed of psbE, psbL and psbJ. The first gene, psbE encodes the α subunits of a transmembrane component of the photosystem II complex [1, 2]. It has also been shown to be closely liked to the psbF gene [3]. Whilst transcription of the psbE gene has been shown to be independent of any other genes, it has been shown that it can be co-transcribed with the psbL and psbJ genes which are located further downstream [4].
The psbL gene is the next gene that forms the psbELJ complex. The complex spans the membrane and contains an alpha helical domain [5]. This domain has a role in supporting the quinone acceptor within the reaction centre's core complex [6] with the carboxyl terminal at the end of the protein responsible for the recovery of electron transfer activity in the complex [7, 8].
The third gene of the psbELJ complex, psbJ forms an entrance for a quinone channel, allowing access to sites within photosystem I [9]. It is thought to be involved in the regulation of electron transfer between the quinone a and the plastoquinone pool [10, 11]. It contains a transmembrane helix [12] and produces a protein which is responsible for the regulation of photosystem II complexes within the thylakoid membrane [13]. It has been indicated that the studies of mutants have shown that organisms without the psbJ gene produce lower electron transfer efficiency in the photosystem II complex [14].
The second operon begins with psbT followed by psbB. Parts psbT and psbB make up the light harvesting complex inside the photosystem II complex [15]. They have a close interaction with the psbA and psbD complex helping to provide excitation energy from outer complexes onto them [16]. The psbT protein is a hydrophobic peptide that also has a single transmembrane helix which is associated with the PSII core complex [17] which binds to the complexes D1 and D2 heterodimer [18]. It's location in the core complex is believe to be at the PSII dimer interface [19]. Mutations in the psbT gene indicate that it's expression is not necessarily essential for the function of the PSII complex [18]. The role of psbT in repair of photo-damaged PSII complexes has also been discussed [17]. In addition the psbT gene maintains the stability of QA [20].
The final gene of the composite part is psbB. The psbB protein is associated with the thylakoid membrane and forms part of the inner light harvesting complexes of PSII [21, 22].
Part Verification

Fig 1. Gel electrophoresis of the operons and single parts constituting the Photosystem II pathway implemented in this project. The part psbELJ-TB (2118 bp) can be seen in lane 8 on the gel at the correct size relative to the 1kb ladder and other parts in our project. The band lower down in this lane on the gel is the pSB1C3 backbone.
Protein information
psbE
mass: 9.3kDa
sequence: MAGKPVERPFSDILTSIRYWVIHSITVPALFIAGWLFVSTGLAYDVFGTPRPNEYFTEDRQEAPLITDRFNALEQVKKLSGN
psbL
mass: 4.43kDa
sequence: MARPNPNKQVVELNRTSLYWGLLLIFVLAVLFSSYIFN
psbJ
mass: 4.29kDa
sequence: MSNTGTTGRIPLWLVGTVVGTLAIGLLAVFFYGSYVGLGSSL
psbT
mass: 3.64kDa
sequence: MEALVYTFLLVGTLGIIFFSIFFRDPPRMIK
psbB
mass: 56.11kDa
sequence: MGLPWYRVHTVVINDPGRLISVHLMHTALVSGWAGSMALFEISVFDPSDPVLNPMWRQGMFVLPFMTRLGITQSWGGWTISGETATNPGIWSYEGVAAAHIILSGALFLASVWHWTYWDLELFRDPRTGKTALDLPKIFGIHLFLSGLLCFGFGAFHVTGVFGPGIWVSDPYGLTGRVQPVAPSWGADGFDPYNPGGIASHHIAAGILGVLAGLFHLCVRPSIRLYFGLSMGSIETVLSSSIAAVFWAAFVVAGTMWYGSAATPIELFGPTRYQWDQGFFQQEIQKRVQASLAEGASLSDAWSRIPEKLAFYDYIGNNPAKGGLFRTGAMNSGDGIAVGWLGHASFKDQEGRELFVRRMPTFFETFPVLLLDKDGIVRADVPFRKAESKYSIEQVGVSVTFYGGELDGLTFTDPATVKKYARKAQLGEIFEFDRSTLQSDGVFRSSPRGWFTFGHVCFALLFFFGHIWHGARTIFRDVFAGIDDDINDQVEFGKYKKLGDTSSLREAF
References
[1] Cramer WA, Tae GS, Furbacher PN, Böttger M. The enigmatic cytochrome b‐559 of oxygenic photosynthesis. Physiologia Plantarum. 1993 Aug 1;88(4):705-11.
[2] Tae GS, Black MT, Cramer WA, Vallon O, Bogorad L. Thylakoid membrane protein topography: transmembrane orientation of the chloroplast cytochrome b-559 psbE gene product. Biochemistry. 1988 Dec;27(26):9075-80.
[3] Erickson JM, Rochaix JD. The photosystems: Structure, function, and molecular biology. Topics in Photosynthesis. 1992;11:101-78.
[4] Kiss É, Kós PB, Chen M, Vass I. A unique regulation of the expression of the psbA, psbD, and psbE genes, encoding the D1, D2 and cytochrome b559 subunits of the Photosystem II complex in the chlorophyll d containing cyanobacterium Acaryochloris marina. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 2012 Jul 31;1817(7):1083-94.
[5] Anbudurai PR, Pakrasi HB. Mutational analysis of the psbL protein of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Zeitschrift für Naturforschung C. 1993 Apr 1;48(3-4):267-74.
[6] Kitamura K, Ozawa S, Shiina T, Toyoshima Y. L protein, encoded by psbL, restores normal functioning of the primary quinone acceptor, QA, in isolated D1/D2/CP47/Cytb‐559/I photosystem II reaction center core complex. FEBS letters. 1994 Oct 31;354(1):113-6.
[7] Ozawa S, Kobayashi T, Sugiyama R, Hoshida H, Shiina T, Toyoshima Y. Role of PSII-L protein (psbL gene product) on the electron transfer in photosystem II complex. 1. Over-production of wild-type and mutant versions of PSII-L protein and reconstitution into the PSII core complex. Plant molecular biology. 1997 May 1;34(1):151-61.
[8] Suorsa M, Regel RE, Paakkarinen V, Battchikova N, Herrmann RG, Aro EM. Protein assembly of photosystem II and accumulation of subcomplexes in the absence of low molecular mass subunits PsbL and PsbJ. European Journal of Biochemistry. 2004 Jan 1;271(1):96-107.
[9] Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nature structural & molecular biology. 2009 Mar 1;16(3):334-42.
[10] Regel RE, Ivleva NB, Zer H, Meurer J, Shestakov SV, Herrmann RG, Pakrasi HB, Ohad I. Deregulation of electron flow within photosystem II in the absence of the psbJ protein. Journal of Biological Chemistry. 2001 Nov 2;276(44):41473-8.
[11] Lind LK, Shukla VK, Nyhus KJ, Pakrasi HB. Genetic and immunological analyses of the cyanobacterium Synechocystis sp. PCC 6803 show that the protein encoded by the psbJ gene regulates the number of photosystem II centers in thylakoid membranes. Journal of Biological Chemistry. 1993 Jan 25;268(3):1575-9.
[12] Nowaczyk MM, Krause K, Mieseler M, Sczibilanski A, Ikeuchi M, Rögner M. Deletion of psbJ leads to accumulation of Psb27–Psb28 photosystem II complexes in Thermosynechococcus elongatus. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 2012 Aug 31;1817(8):1339-45.
[13] Lind LK, Shukla VK, Nyhus KJ, Pakrasi HB. Genetic and immunological analyses of the cyanobacterium Synechocystis sp. PCC 6803 show that the protein encoded by the psbJ gene regulates the number of photosystem II centers in thylakoid membranes. Journal of Biological Chemistry. 1993 Jan 25;268(3):1575-9.
[14] Regel RE, Ivleva NB, Zer H, Meurer J, Shestakov SV, Herrmann RG, Pakrasi HB, Ohad I. Deregulation of electron flow within photosystem II in the absence of the psbJ protein. Journal of Biological Chemistry. 2001 Nov 2;276(44):41473-8.
[15] Barber J. Photosystem two. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 1998 Jun 10;1365(1):269-77.
[16] Luciński R, Jackowski G. The structure, functions and degradation of pigment-binding proteins of photosystem II. Acta Biochim Pol. 2006;53(4):693-708.
[17] Monod C, Takahashi Y, Goldschmidt-Clermont M, Rochaix JD. The chloroplast ycf8 open reading frame encodes a photosystem II polypeptide which maintains photosynthetic activity under adverse growth conditions. The EMBO journal. 1994 Jun 15;13(12):2747.
[18]Ohnishi N, Takahashi Y. psbT polypeptide is required for efficient repair of photodamaged photosystem II reaction center. Journal of Biological Chemistry. 2001 Sep 7;276(36):33798-804.
[19] Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004 Mar 19;303(5665):1831-8.
[20] Ohnishi N, Kashino Y, Satoh K, Ozawa SI, Takahashi Y. Chloroplast-encoded polypeptide psbT is involved in the repair of primary electron acceptor QA of photosystem II during photoinhibition in Chlamydomonas reinhardtii. Journal of Biological Chemistry. 2007 Mar 9;282(10):7107-15.
[21] Barber J, Nield J, Morris EP, Zheleva D, Hankamer B. The structure, function and dynamics of photosystem two. Physiologia plantarum. 1997 Aug 1;100(4):817-27.
[22] Bricker TM, Frankel LK. The structure and function of CP47 and CP43 in photosystem II. Photosynthesis research. 2002 May 1;72(2):131-46.
| None |
